The amount of experimentally accumulated data about DNA, RNA, and protein sequences is overwhelming, but we still do not know the functions of the majority genes in the human genome. The theoretical quantititative methods from statistical physics prove to be very useful in this area. We have successfully applied these methods to the following problems:
A KRAB/KAP1-miRNA Cascade Regulates Erythropoiesis Through Stage-Specific Control of Mitophagy
Based on our predictions, Didier Trono’s group at EPFL was able to discover and experimentally confirm the role of microRNA in the maturation of red blood cells (Barde et al 2013)
Prediction of microRNA targets
You can access the microRNA target predictions of different versions of our algorithm here.
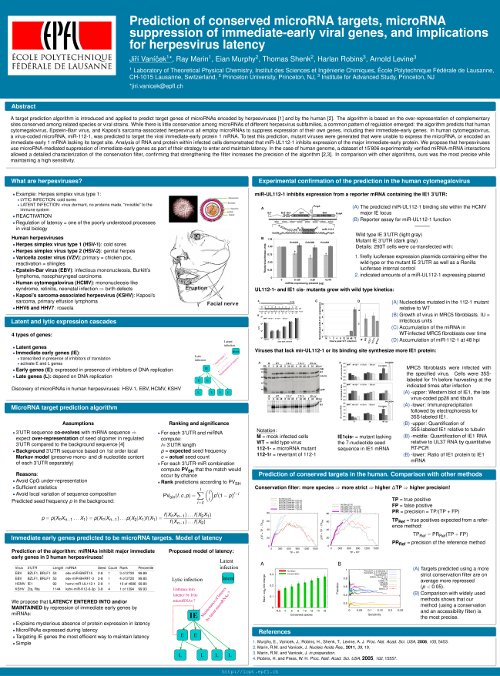
1) Prediction of microRNA targets in herpesviruses and implications for herpesvirus latency
Published in Proceedings of the National Academy of Sciences USA, 105, p 5453-5458 (2008)
We have developed an algorithm to predict microRNA targets, based on overrepresentation (originally used by Robins and Press, PNAS 2005) of conserved potential binding sites in the 3’UTR of genes of interest. Our predictions were confirmed experimentally on human cytomegalovirus by Eain Murphy and Tom Shenk from Princeton. The results are published in a PNAS article (Eain et al 2008).
Patent application “MicroRNAs for Prevention and Therapy of Herpes Virus Disease” has been filed in the United States Patent and Trademark Office.
| Download the PDF version of the poster |
 |
| Download the PDF version of the poster |
2) Efficient use of accessibility of potential binding sites in microRNA target prediction
Published in Nucleic Acids Research, 39, p 19-29 (2011)
→ Access PACMIT predictions in the Human and in Drosophila malanogaster
We have developed a version of our target prediction algorithm that allows considering the secondary structure of the target sequence. By including the accessibility of the binding site in the algorithm we reduced the rate of false positives in the predictions. This algorithm is called PACMIT (Prediction of ACcessible MIcroRNA Targets) (Marin and Vanicek 2011).
| Download the PDF version of the poster |
 |
| Download the PDF version of the poster |
3) Taking advantage of combined conservation and accessibility filters in microRNA target prediction
Published in PLoS ONE, 7, e32208, (2012)
→ Access PACCMIT predictions in the Human
We have developed a version of our algorithm that can use both conservation and accessibility filters simultaneously. The new and general version, called PACCMIT (Prediction of ACcessible and Conserved MIcroRNA Targets), leads to a further increase in precision of target predictions (Marin and Vanicek 2012).
Detection of functional RNA and DNA signals in the human genome
In collaboration with Todd Riley, Michael Krasnitz, and Arnold Levine from the Institute for Advanced Study.